Former group Prof. Dr. Frasch
Drosophila Mesoderm Development
Patterning and Organogenesis in the Drosophila mesoderm
Our lab is studying the processes of mesoderm patterning and mesodermal tissue development in Drosophila. Genetic and molecular studies aim to define the identities and functions of regulatory molecules that spatially subdivide the mesodermal cell layer and determine the primordial cells of the heart, skeletal, and visceral musculatures at defined locations in the early embryo. We are also interested in processes of cell migration, tissue morphogenesis, and plasticity that are important for normal muscle and heart development.

Combinatorial signals and transcriptional inputs during progenitor specification
We have shown that several of the identified mesoderm patterning processes involve synergistic activities of mesoderm-specific transcription factors and signaling molecules that are secreted from the ectoderm. For example, the mesodermally-expressed homeobox gene tinman cooperates with the ectodermally-secreted factor Dpp, a TGF-beta molecule of the BMP family, to subdivide the mesoderm into a ventral and a dorsal portion and to determine dorsal mesodermal derivatives. Molecularly, this process involves the binding of Dpp-activated signal transducers (the Smad proteins Mad and Medea), together with the Tinman protein itself, to target enhancers of genes such as tinman, bagpipe, and even-skipped, which results in their spatially-restricted activation in the dorsal mesoderm. Other spatial cues such as Wingless and the forkhead domain protein Sloppy Paired provide additional inputs for subdividing the dorsal mesoderm into heart, gut muscle, and somatic muscle primordia. We are interested in finding out how these combinatorial activities cooperate at the molecular level to activate or repress developmental control genes in defined groups of cells. Moreover, we study how some of these control genes (e.g., the Dorsocross and org-1 T-box genes, the homeobox gene bagpipe, the forkhead domain encoding gene biniou, and others) ultimately specify cell fates of the cardiac, visceral, and skeletal musculatures. We also perform genetic and genomic screens to identify new targets genes of these control genes as well as new regulators of mesodermal tissue development.
Representative Publications
Jin, H., Stojnic, R., Adryan, B., Ozdemir, A., Stathopoulos, A., Frasch, M. (2013) Genome-wide screens for in vivo Tinman binding sites identify cardiac enhancers with diverse functional architectures. PLoS Genet 9, e1003195.
Schaub, C., Nagaso, H., Jin, H., Frasch, M. (2012) Org-1, the Drosophila ortholog of Tbx1, is a direct activator of known identity genes during muscle specification. Development 139, 1001-1012.
Lee, H. H., Norris, A., Weiss, J. B., Frasch, M. (2003) Jelly belly protein activates the receptor tyrosine kinase Alk to specify visceral muscle pioneers. Nature 425, 507-512.
Xu, X., Yin, Z., Hudson, J., Ferguson, E., Frasch, M. (1998) Smad proteins act in combination with synergistic and antagonistic regulators to target Dpp responses to the Drosophila mesoderm. Genes Dev 12, 2354-2370.
Frasch, M. (1995) Induction of visceral and cardiac mesoderm by ectodermal Dpp in the early Drosophila embryo. Nature 374, 464-467.
Cell migration and muscle morphogenesis
Prior to their fusion with fusion-competent myoblasts, the founder cells of the somatic muscles and particularly the longitudinal gut muscles undergo precisely-controlled cell migrations. We are currently aiming to identify some of the guidance cues and other mechanisms that are essential for the migration of these cells to their destinations.
Representative Publications
Reim, I., Hollfelder, D., Ismat, A., Frasch, M. (2012) The FGF8-related signals Pyramus and Thisbe promote pathfinding, substrate adhesion, and survival of migrating longitudinal gut muscle founder cells. Dev Biol 368, 28-43.
Ismat, A., Schaub, C., Reim, I., Kirchner, K., Schultheis, D., Frasch, M. (2010) HLH54F is required for the specification and migration of longitudinal gut muscle founders from the caudal mesoderm of Drosophila. Development 137, 3107-3117.
Syncytial muscle dedifferentiation and transdifferentiation
We have identified some striking examples in which syncytial muscles dedifferentiate, fragment into mononucleated myoblasts, and subsequently redifferentiate into the same or transdifferentiate into different types of syncytial muscles during development. One example is the transdifferentiation of certain alary muscles into heart-associated ventral longitudinal muscles during metamorphosis. We are interested in defining the molecular mechanisms executing these unusual processes of muscle plasticity.
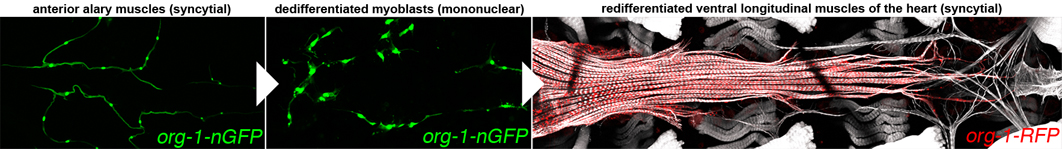
Representative Publication
Schaub, C., März, J., Reim, I., Frasch, M. (2015) Org-1-Dependent Lineage Reprogramming Generates the Ventral Longitudinal Musculature of the Drosophila Heart. Curr Biol 25, 488-494.
Evolutionary conservation of muscle and heart development
Importantly, most of the identified regulators and pathways in mesoderm development are evolutionarily conserved. It appears that mesoderm patterning, muscle development, and cardiogenesis in insect and vertebrate embryos involve many closely related mechanisms. Therefore, Drosophila can serve as an excellent model system for developmental processes of the heart, skeletal and gut muscles, as well as organogenesis of other internal organs, in vertebrate embryos. We are taking further advantage of evolutionary conservation in muscle development by screening for genes that are required for somatic muscle development in the beetle Tribolium, which provides the benefit of systemic RNA interference. Within the DFG-funded iBeetle consortium, a genome-wide RNAi screen has been performed, in which muscle-GFP-marked embryos are screened for muscle defects upon pupal double-stranded RNA injections. The functions of newly-identified regulatory genes and their orthologs are being analyzed both in Tribolium and in Drosophila.
Representative Publication
Schmitt-Engel, C., Schultheis, D., Schwirz, J., Ströhlein, N., Troelenberg, N., Majumdar, U., … Frasch, M., Roth, S., Wimmer, E. A., Schoppmeier, M., Klingler, M., Bucher, G. (2015) The iBeetle large-scale RNAi screen reveals gene functions for insect development and physiology. Nat Commun 6, 7822.
Members
Claudia Obermeier (BTA)

Lab alumni
| Name | Position | Period | Current Position |
| Cord Dohrmann | Diploma Student | 1990-1991 | Chief Scientific Officer, Evotec AG, Hamburg |
| Natalia Azpiazu | Graduate Student | 1991-1994 | Group Leader CBM/Madrid University, Spain |
| Polonca Küssel | Graduate Student | 1990-1995 | Director, Head of Europe Global Clinical Operations, Teva Pharmaceuticals |
| Xiaolei Xu | Graduate Student | 1994-1999 | Associate Professor, Mayo Clinic, Rochester MI |
| Stefan Knirr | Postdoc | 1997-2000 | Patent Attorney, Research Foundation of CUNY & Adjunct Associate Prof., Hunter College New York, NY |
| Stephane Zaffran | Postdoc | 1997-2001 | Directeur de recherche, Inserm UMR 910, Faculte de Medecine La Timone, Marseilles, FR |
| Hsiu-Hsiang Lee | Graduate Student
Postdoc |
1997-2003
2003-2004 |
Assistant Professor, National Taiwan University, Taipeh |
| Patrick Lo | Postdoc, Associate | 1994-2007 | Associate Editor, BioTechniques |
| Hideyuki Nagaso | Postdoc | 2001-2007 | Pharma business, Hokkaido, JP |
| Ingolf Reim | Postdoc | 2001-2007 | Group Leader, University of Erlangen-Nuremberg, Developmental Biology |
| Afshan Ismat | Graduate Student | 2003-2007 | Assistant Professor, University of St. Thomas, St. Paul, MN |
| Dominik Müller | Postdoc | 2005-2009 | High School teacher, Helene-Lang-Gymnasium, Fürth |
| Hong Jin | Graduate Student | 2008-2012 | Postdoc, Department of Biochemistry & Molecular Biology, University of Calgary, Canada |
| Dorothea Schultheis | Graduate Student | 2010-
2016 |
Postdoc, FAU Erlangen-Nürnberg, Dept. of Medicine, Div. of Neuropathology, Muscle Lab |
